The Power of Optimization in Designing Experiments Involving Small Samples
A step-by-step guide to designing more precise experiments using optimization in PythonExperimentation is the foundation for testing hypotheses with scientific rigor. In medicine, it is used to assess the effect of new treatments on patients, while in the digital world, tech giants like Amazon, Netflix, and Uber run thousands of experiments each year to optimize and improve their platforms.For large-scale experiments, random assignment is commonly used and its considered the “Gold Standard”. With a substantial amount of data, randomness tends to produce comparable groups, where important pre-experiment factors are balanced and the exchangeability assumption holds.However, when the sample for an experiment is very small, random assignment often fails to create statistically equivalent groups. So, how can we split units efficiently between treatment and control groups?What you will learn:In this post, I’ll explain an optimization-based approach to constructing equivalent groups for an experiment which was proposed by Bertsimas et al. in this article.With a simple example in Python we’ll learn how to:Design an optimization-based experiment.Perform inference using bootstrap techniques.Implement the code in Python for your own experimentsThe Power of OptimizationBefore diving into our example and Python code, let’s first discuss some insights into the benefits of using an optimization-based approach to design experiments.Optimization makes statistics more precise, allowing for a more powerful inference.The optimization-based approach matches the experimental groups to minimize the en-masse discrepancies in means and variances. This makes the statistics much more precise, concentrating them tightly around their nominal values while still being unbiased estimates. This increased precision allows for more powerful inference (using the bootstrap algorithm, which we’ll cover later).This allows researchers to draw statistically valid conclusions with less data, reducing experimental costs — an important benefit in disciplines like oncology research, where testing chemotherapy agents in mouse cancer models is both laborious and expensive. Moreover, compared to other methods for small sample sizes, optimization has been shown to outperform them, as we’ll see later with simulated experiments.Now, let’s dive into an example to see how to apply this approach using Python!Implementing the Algorithm in PythonCase Study: A Drug Experiment with 20 MiceLet’s consider an experiment with 20 mice, where we are interested in studying the effect of a drug on tumor growth with varying initial tumor sizes. Suppose that the initial tumor sizes are normally distributed with a mean of 200 mg and a standard deviation of 300 mg (truncated to ensure non-negative values). We can generate the population of mice with the following Python code:import numpy as npimport pandas as pdimport scipy.stats as statsdef create_experiment_data(n_mice, mu, sigma, seed): lower, upper = 0, np.inf initial_weights = stats.truncnorm( (lower - mu) / sigma, (upper - mu) / sigma, loc=mu, scale=sigma ).rvs(size=n_mice, random_state=seed) return pd.DataFrame({ 'mice': list(range(1, n_mice+1)), 'initial_weight': initial_weights, })tumor_data = create_experiment_data(n_mice=20, mu=200, sigma=300, seed=123)print(tumor_data.head())> mice initial_weight0 1 424.7368881 2 174.6910352 3 141.0164783 4 327.5187494 5 442.239789Now, we need to divide the 20 rodents into two groups of 10 each — one group will receive the treatment, while the other will receive a placebo. We’ll accomplish this using optimization.We will also assume that tumor growth is observed over a one-day period and follows the Gompertz model (detailed in Mathematical Models in Cancer Research). The treatment is assumed to have a deterministic effect of reducing tumor size by 250 mg.Experimental DesignWe aim to formulate the creation of equivalent groups as an optimization problem, where the objective is to minimize the discrepancy in both the mean and variance of the initial tumor weight.To implement this, we need to follow three steps:Paso 1: Normalize the Initial Tumor WeightFirst, the entire sample must be pre-processed and the metric should be normalized so that it has a mean of zero and unit variance:mean = tumor_data['initial_weight'].mean()std = tumor_data['initial_weight'].std()tumor_data['norm_initial_weight'] = (tumor_data['initial_weight'] - mean) / stdPaso 2: Create the groups using optimizationNext, we need to implement the general optimization model that construct m-groups with k-subjects each, minimizing the maximum discrepancy between any two groups (a full description of the model variables can be found in the article) and passing it the normalized metric:Optimization model that creates m-groups of k-units each (from Bertsimas et al. 2015).The mathematical model can
A step-by-step guide to designing more precise experiments using optimization in Python
Experimentation is the foundation for testing hypotheses with scientific rigor. In medicine, it is used to assess the effect of new treatments on patients, while in the digital world, tech giants like Amazon, Netflix, and Uber run thousands of experiments each year to optimize and improve their platforms.
For large-scale experiments, random assignment is commonly used and its considered the “Gold Standard”. With a substantial amount of data, randomness tends to produce comparable groups, where important pre-experiment factors are balanced and the exchangeability assumption holds.
However, when the sample for an experiment is very small, random assignment often fails to create statistically equivalent groups. So, how can we split units efficiently between treatment and control groups?
What you will learn:
In this post, I’ll explain an optimization-based approach to constructing equivalent groups for an experiment which was proposed by Bertsimas et al. in this article.
With a simple example in Python we’ll learn how to:
- Design an optimization-based experiment.
- Perform inference using bootstrap techniques.
- Implement the code in Python for your own experiments
The Power of Optimization
Before diving into our example and Python code, let’s first discuss some insights into the benefits of using an optimization-based approach to design experiments.
Optimization makes statistics more precise, allowing for a more powerful inference.
The optimization-based approach matches the experimental groups to minimize the en-masse discrepancies in means and variances. This makes the statistics much more precise, concentrating them tightly around their nominal values while still being unbiased estimates. This increased precision allows for more powerful inference (using the bootstrap algorithm, which we’ll cover later).
This allows researchers to draw statistically valid conclusions with less data, reducing experimental costs — an important benefit in disciplines like oncology research, where testing chemotherapy agents in mouse cancer models is both laborious and expensive. Moreover, compared to other methods for small sample sizes, optimization has been shown to outperform them, as we’ll see later with simulated experiments.
Now, let’s dive into an example to see how to apply this approach using Python!
Implementing the Algorithm in Python
Case Study: A Drug Experiment with 20 Mice
Let’s consider an experiment with 20 mice, where we are interested in studying the effect of a drug on tumor growth with varying initial tumor sizes. Suppose that the initial tumor sizes are normally distributed with a mean of 200 mg and a standard deviation of 300 mg (truncated to ensure non-negative values). We can generate the population of mice with the following Python code:
import numpy as np
import pandas as pd
import scipy.stats as stats
def create_experiment_data(n_mice, mu, sigma, seed):
lower, upper = 0, np.inf
initial_weights = stats.truncnorm(
(lower - mu) / sigma,
(upper - mu) / sigma,
loc=mu,
scale=sigma
).rvs(size=n_mice, random_state=seed)
return pd.DataFrame({
'mice': list(range(1, n_mice+1)),
'initial_weight': initial_weights,
})
tumor_data = create_experiment_data(n_mice=20, mu=200, sigma=300, seed=123)
print(tumor_data.head())
> mice initial_weight
0 1 424.736888
1 2 174.691035
2 3 141.016478
3 4 327.518749
4 5 442.239789
Now, we need to divide the 20 rodents into two groups of 10 each — one group will receive the treatment, while the other will receive a placebo. We’ll accomplish this using optimization.
We will also assume that tumor growth is observed over a one-day period and follows the Gompertz model (detailed in Mathematical Models in Cancer Research). The treatment is assumed to have a deterministic effect of reducing tumor size by 250 mg.
Experimental Design
We aim to formulate the creation of equivalent groups as an optimization problem, where the objective is to minimize the discrepancy in both the mean and variance of the initial tumor weight.
To implement this, we need to follow three steps:
Paso 1: Normalize the Initial Tumor Weight
First, the entire sample must be pre-processed and the metric should be normalized so that it has a mean of zero and unit variance:
mean = tumor_data['initial_weight'].mean()
std = tumor_data['initial_weight'].std()
tumor_data['norm_initial_weight'] = (tumor_data['initial_weight'] - mean) / std
Paso 2: Create the groups using optimization
Next, we need to implement the general optimization model that construct m-groups with k-subjects each, minimizing the maximum discrepancy between any two groups (a full description of the model variables can be found in the article) and passing it the normalized metric:
The mathematical model can be implemented in Python using the ortools library with the SCIP solver, as follows:
from ortools.linear_solver import pywraplp
from typing import Union
class SingleFeatureOptimizationModel:
"""
Implements the discrete optimization model proposed by Bertsimas et al. (2015) in "The Power of
Optimization Over Randomization in Designing Experiments Involving Small Samples".
See: https://doi.org/10.1287/opre.2015.1361.
"""
def __init__(self, data: pd.DataFrame, n_groups: int, units_per_group: int, metric: str, unit: str):
self.data = data.reset_index(drop=True)
self.parameters = {
'rho': 0.5,
'groups': range(n_groups),
'units': range(len(self.data)),
'unit_list': self.data[unit].tolist(),
'metric_list': self.data[metric].tolist(),
'units_per_group': units_per_group,
}
self._create_solver()
self._add_vars()
self._add_constraints()
self._set_objective()
def _create_solver(self):
self.model = pywraplp.Solver.CreateSolver("SCIP")
if self.model is None:
raise Exception("Failed to create SCIP solver")
def _add_vars(self):
self.d = self.model.NumVar(0, self.model.infinity(), "d")
self.x = {}
for i in self.parameters['units']:
for p in self.parameters['groups']:
self.x[i, p] = self.model.IntVar(0, 1, "")
def _set_objective(self):
self.model.Minimize(self.d)
def _add_constraints(self):
self._add_constraints_d_bounding()
self._add_constraint_group_size()
self._add_constraint_all_units_assigned()
def _add_constraints_d_bounding(self):
rho = self.parameters['rho']
for p in self.parameters['groups']:
for q in self.parameters['groups']:
if p < q:
self.model.Add(self.d >= self._mu(p) - self._mu(q) + rho * self._var(p) - rho * self._var(q))
self.model.Add(self.d >= self._mu(p) - self._mu(q) + rho * self._var(q) - rho * self._var(p))
self.model.Add(self.d >= self._mu(q) - self._mu(p) + rho * self._var(p) - rho * self._var(q))
self.model.Add(self.d >= self._mu(q) - self._mu(p) + rho * self._var(q) - rho * self._var(p))
def _add_constraint_group_size(self):
for p in self.parameters['groups']:
self.model.Add(
self.model.Sum([
self.x[i,p] for i in self.parameters['units']
]) == self.parameters['units_per_group']
)
def _add_constraint_all_units_assigned(self):
for i in self.parameters['units']:
self.model.Add(
self.model.Sum([
self.x[i,p] for p in self.parameters['groups']
]) == 1
)
def _add_contraint_symmetry(self):
for i in self.parameters['units']:
for p in self.parameters['units']:
if i < p:
self.model.Add(
self.x[i,p] == 0
)
def _mu(self, p):
mu = self.model.Sum([
(self.x[i,p] * self.parameters['metric_list'][i]) / self.parameters['units_per_group']
for i in self.parameters['units']
])
return mu
def _var(self, p):
var = self.model.Sum([
(self.x[i,p]*(self.parameters['metric_list'][i])**2) / self.parameters['units_per_group']
for i in self.parameters['units']
])
return var
def optimize(
self,
max_run_time: int = 60,
max_solution_gap: float = 0.05,
max_solutions: Union[int, None] = None,
num_threads: int = -1,
verbose: bool = False
):
"""
Runs the optimization model.
Args:
max_run_time: int
Maximum run time in minutes.
max_solution_gap: float
Maximum gap with the LP relaxation solution.
max_solutions: int
Maximum number of solutions until stop.
num_threads: int
Number of threads to use in solver.
verbose: bool
Whether to set the solver output.
Returns: str
The status of the solution.
"""
self.model.SetTimeLimit(max_run_time * 60 * 1000)
self.model.SetNumThreads(num_threads)
if verbose:
self.model.EnableOutput()
self.model.SetSolverSpecificParametersAsString(f"limits/gap = {max_solution_gap}")
self.model.SetSolverSpecificParametersAsString(f"limits/time = {max_run_time * 60}")
if max_solutions:
self.model.SetSolverSpecificParametersAsString(f"limits/solutions = {max_solutions}")
status = self.model.Solve()
if verbose:
if status == pywraplp.Solver.OPTIMAL:
print("Optimal Solution Found.")
elif status == pywraplp.Solver.FEASIBLE:
print("Feasible Solution Found.")
else:
print("Problem infeasible or unbounded.")
self._extract_solution()
return status
def _extract_solution(self):
tol = 0.01
self.assignment = {}
for i in self.parameters['units']:
for p in self.parameters['groups']:
if self.x[i,p].solution_value() > tol:
self.assignment.setdefault(p, []).append(self.parameters['unit_list'][i])
def get_groups_list(self):
return list(self.assignment.values())
model = SingleFeatureOptimizationModel(
data = tumor_data,
n_groups = 2,
units_per_group = 10,
unit = 'mice',
metric = 'norm_initial_weight',
)
status = model.optimize()
optimized_groups = model.get_groups_list()
print(f"The optimized mice groups are: {optimized_groups}")
> The optimized mice groups are: [
[1, 4, 5, 6, 8, 12, 14, 16, 17, 18],
[2, 3, 7, 9, 10, 11, 13, 15, 19, 20]
]
Note: The parameter rho controls the trade-off between minimizing discrepancies in the first moment and second moment and is chosen by the researcher. In our example, we have considered rho equals 0.5.
Paso 3: Randomize which group receives which treatment
Lastly, we randomly determine which experimental mice group will receive the drug, and which will receive the placebo:
import random
def random_shuffle_groups(group_list, seed):
random.seed(seed)
random.shuffle(group_list)
return group_list
randomized_groups = random_shuffle_groups(optimized_groups, seed=123)
treatment_labels = ["Placebo", "Treatment"]
treatment_dict = {treatment_labels[i]: randomized_groups[i] for i in range(len(randomized_groups))}
print(f"The treatment assignment is: {treatment_dict}")
> The treatment assignment is: {
'Placebo': [2, 3, 7, 9, 10, 11, 13, 15, 19, 20],
'Treatment': [1, 4, 5, 6, 8, 12, 14, 16, 17, 18]
}Let’s see the quality of the result by analyzing the mean and variance of the initial tumor weights in both groups:
mice_assignment_dict = {inx: gp for gp, indices in treatment_dict.items() for inx in indices}
tumor_data['treatment'] = tumor_data['mice'].map(mice_assignment_dict)
print(tumor_data.groupby('treatment').agg(
avg_initial_weight = ('initial_weight', 'mean'),
std_initial_weight = ('initial_weight', 'std'),
).round(2))> avg_initial_weight std_initial_weight
treatment
Placebo 302.79 202.54
Treatment 303.61 162.12
This is the group division with minimal discrepancy in the mean and variance of pre-experimental tumor weights! Now let’s conduct the experiment and analyze the results.
Inference with bootstrap
Simulating tumor growth
Given the established treatment assignment, tumor growth is simulated over one day using the Gompertz model, assuming an effect of -250 mg for the treated group:
import numpy as np
from scipy.integrate import odeint
# Gompertz model parameters
a = 1
b = 5
# Critical weight
wc = 400
def gompex_growth(w, t):
"""
Gomp-ex differential equation model based on the initial weight.
"""
growth_rate = a + max(0, b * np.log(wc / w))
return w * growth_rate
def simulate_growth(w_initial, t_span):
"""
Simulate the tumor growth using the Gomp-ex model.
"""
return odeint(gompex_growth, w_initial, t_span).flatten()
def simulate_tumor_growth(data: pd.DataFrame, initial_weight: str, treatment_col: str, treatment_value: str, treatment_effect: float):
"""
Simulate the tumor growth experiment and return the dataset.
"""
t_span = np.linspace(0, 1, 2)
final_weights = np.array([simulate_growth(w, t_span)[-1] for w in data[initial_weight]])
experiment_data = data.copy()
mask_treatment = data[treatment_col] == treatment_value
experiment_data['final_weight'] = np.where(mask_treatment, final_weights + treatment_effect, final_weights)
return experiment_data.round(2)
experiment_data = simulate_tumor_growth(
data = tumor_data,
initial_weight = 'initial_weight',
treatment_col = 'treatment',
treatment_value = 'Treatment',
treatment_effect = -250
)
print(experiment_data.head())
> mice initial_weight norm_initial_weight treatment final_weight
0 1 424.74 0.68 Treatment 904.55
1 2 174.69 -0.72 Placebo 783.65
2 3 141.02 -0.91 Placebo 754.56
3 4 327.52 0.14 Treatment 696.60
4 5 442.24 0.78 Treatment 952.13
mask_tr = experiment_data.group == 'Treatment'
mean_tr = experiment_data[mask_tr]['final_weight'].mean()
mean_co = experiment_data[~mask_tr]['final_weight'].mean()
print(f"Mean difference between treatment and control: {round(mean_tr - mean_co)} mg")
> Mean difference between treatment and control: -260 mg
Now that we have the final tumor weights, we observe that the average final tumor weight in the treatment group is 260 mg lower than in the control group. However, to determine if this difference is statistically significant, we need to apply the following bootstrap mechanism to calculate the p-value.
Bootstrap inference for optimization-based design
“In an optimization-based design, statistics like the average difference between groups become more precise but no longer follow the usual distributions. Therefore, a bootstrap inference method should be used to draw valid conclusions.”
The boostrap inference method proposed by Bertsimas et al. (2015) involves using sampling with replacement to construct the baseline distribution of our estimator. In each iteration, the group division is performed using optimization, and finally the the p-value is derived as follows:
from tqdm import tqdm
from typing import Any, List
def inference(data: pd.DataFrame, unit: str, outcome: str, treatment: str, treatment_value: Any = 1, n_bootstrap: int = 1000) -> pd.DataFrame:
"""
Estimates the p-value using bootstrap for two groups.
Parameters
-----------
data (pd.DataFrame): The experimental dataset with the observed outcome.
unit (str): The experimental unit column.
outcome (str): The outcome metric column.
treatment (str): The treatment column.
treatment_value (Any): The value referencing the treatment (other will be considered as control).
n_bootstrap (int): The number of random draws with replacement to use.
Returns
-----------
pd.DataFrame: The dataset with the results.
Raise
------------
ValueException: if there are more than two treatment values.
"""
responses = data[outcome].values
mask_tr = (data[treatment] == treatment_value).values
delta_obs = _compute_delta(responses[mask_tr], responses[~mask_tr])
deltas_B = _run_bootstrap(data, unit, outcome, n_bootstrap)
pvalue = _compute_pvalue(delta_obs, deltas_B)
output_data = pd.DataFrame({
'delta': [delta_obs],
'pvalue': [pvalue],
'n_bootstrap': [n_bootstrap],
'avg_delta_bootstrap': [np.round(np.mean(deltas_B), 2)],
'std_delta_bootstrap': [np.round(np.std(deltas_B), 2)]
})
return output_data
def _run_bootstrap(data: pd.DataFrame, unit: str, outcome: str, B: int = 1000) -> List[float]:
"""
Runs the bootstrap method and returns the bootstrapped deltas.
Parameters
-----------
data (pd.DataFrame): The dataframe from which sample with replacement.
outcome (str): The outcome metric observed in the experiment.
B (int): The number of random draws with replacement to perfrom.
Returns
-----------
List[float]: The list of bootstrap deltas.
"""
deltas_bootstrap = []
for i in tqdm(range(B), desc="Bootstrap Progress"):
sample_b = _random_draw_with_replacement(data, unit)
responses_b, mask_tr_b = _optimal_treatment_control_split(sample_b, unit, outcome, seed=i)
delta_b = _compute_delta(responses_b[mask_tr_b], responses_b[~mask_tr_b])
deltas_bootstrap.append(delta_b)
return deltas_bootstrap
def _compute_delta(response_tr, responses_co):
delta = np.mean(response_tr) - np.mean(responses_co)
return delta
def _compute_pvalue(obs_delta, bootstrap_deltas):
count_extreme = sum(1 for delta_b in bootstrap_deltas if abs(delta_b) >= abs(obs_delta))
p_value = (1 + count_extreme) / (1 + len(bootstrap_deltas))
return p_value
def _random_draw_with_replacement(data: pd.DataFrame, unit: str):
sample = data.sample(frac=1, replace=True)
sample[unit] = range(1, len(sample) + 1)
return sample
def _optimal_treatment_control_split(data: pd.DataFrame, unit: str, outcome: str, seed: int):
result = _sample(
data = data,
unit = unit,
normalized_feature = 'norm_initial_weight',
seed = seed
)
treatment_dict = {inx: gp for gp, indices in result.items() for inx in indices}
treatment = data[unit].map(treatment_dict)
mask_tr = (treatment == 'Treatment').values
responses = data[outcome].values
return responses, mask_tr
def _sample(data: pd.DataFrame, unit: str, normalized_feature: str, seed: int):
model = SingleFeatureOptimizationModel(
data,
n_groups = 2,
units_per_group = 10,
unit = unit,
metric = normalized_feature,
)
status = model.optimize()
optimized_groups = model.get_groups_list()
randomized_groups = random_shuffle_groups(optimized_groups, seed=seed)
treatment_labels = ["Placebo", "Treatment"]
return {treatment_labels[i]: randomized_groups[i] for i in range(len(randomized_groups))}
infer_result = inference(
data = experiment_data,
unit = 'mice',
outcome = 'final_weight',
treatment = 'group',
treatment_value = 'Treatment',
n_bootstrap = 1000
)
print(infer_result)
> delta pvalue n_bootstrap avg_delta_bootstrap std_delta_bootstrap
0 -260.183 0.001998 1000 2.02 112.61
The observed difference of -260 mg between the groups is significant at the 5% significance level (the p-value less than 0.05). Therefore, we reject the null hypothesis of equal means and conclude that the treatment had a statistically significant effect.
Results over 1000 mice experiments
We can simulate the experiment multiple times, generating populations of mice with different initial tumor weights, drawn from the same normal distribution with a mean of 200 mg and a standard deviation of 300 mg.
This allows us to compare the optimization-based design with other experimental designs. In the following graph, I compare the optimization approach with simple random assignment and stratified random assignment (where the strata were created using a k-means algorithm based on initial tumor weight):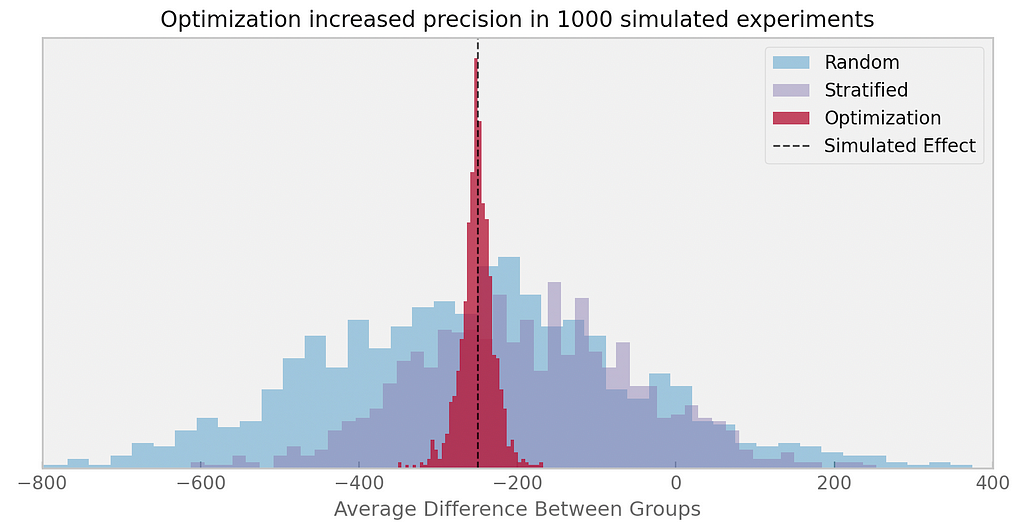
In their article, the authors also compare the optimization-based approach with re-randomization and pairwise matching across different effect sizes and group sizes. I highly recommend reading the full article if you’re interested in exploring the details further!
Congratulations! You’ve reached the end ???? If you found this article interesting, consider following me. I often share ideas about optimization and causal inference.
The Power of Optimization in Designing Experiments Involving Small Samples was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.













